Kanghyun Kim1, Amey Chaware1, Clare B. Cook1, Shiqi Xu1, Monica Abdelmalak2, Colin Cooke3, Kevin C. Zhou1,4, Mark Harfouche5, Paul Reamey5, Veton Saliu5, Jed Doman5, Clay Dugo5, Gregor Horstmeyer2, Richard Davis2, Ian Taylor-Cho2, Wen-Chi Foo2, Lucas Kreiss1, Xiaoyin Sara Jiang2, Roarke Horstmeyer1,2
1Department of Biomedical Engineering, Duke University, Durham, 27708, NC, USA.
2Department of Pathology, Duke University Medical Center, Durham, 27708, NC, USA.
3Department of Electrical and Computer Engineering, Duke University, Durham, 27708, NC, USA.
4Department of Electrical Engineering and Computer Sciences, University of California, Berkeley, 94720, CA, USA.
5Ramona Optics Inc., 1000 W Main St., Durham, NC 27701, USA.
Abstract
Optical microscopy has long been the standard method for diagnosis in cytopathology. Whole slide scanners can image and digitize large sample areas automatically, but are slow, expensive and therefore not widely available. Clinical diagnosis of cytology specimens is especially challenging since these samples are both spread over large areas and thick, which requires 3D capture. Here, we introduce a new parallelized microscope for scanning thick specimens across extremely wide fields-of-view (54×72 mm2) at 1.2 and 0.6 µm resolutions, accompanied by machine learning software to rapidly assess these 16 gigapixel scans. This Multi-Camera Array Scanner (MCAS) comprises 48 micro-cameras closely arranged to simultaneously image different areas. By capturing 624 megapixels per snapshot, the MCAS is significantly faster than most conventional whole slide scanners. We used this system to digitize entire cytology samples (scanning three entire slides in 3D in just several minutes) and demonstrate two machine learning techniques to assist pathologists: first, an adenocarcinoma detection model in lung specimens (0.73 recall); second, a slide-level classification model of lung smears (0.969 AUC).
Overview
Digitizing thick cytology smears is challenging due to the need for high resolutions, the 3D nature of samples, and extremely large resulting datasets, which can be on the order of 100 GB. This has led to a lack of effective solutions for quickly imaging entire smears, hindering cytopathologists from fully utilizing digital pathology benefits. To overcome these challenges and rapidly image thick microscopic samples at high resolution across a large field of view, we introduce a multi-camera array scanner (MCAS). The MCAS comprises 48 individual image sensors organized into a tightly packed (9 mm spacing) 6 X 8 grid. Each camera is outfitted with a custom-designed, high-resolution objective lens capable of resolving sub-cellular features. Additionally, the MCAS is equipped with sample stages and associated novel firmware to rapidly translate up to 3 slides at once to complete 3D scanning. We showcase the MCAS digitizing full, axially thick smears into multi-gigapixel composites at two unique resolutions – 1.2 μm and 0.6 μm full-pitch resolution (comparable with standard 10X and 20X objective lenses) – by scanning across depths up to 150 μm. This system can capture the entire volume of a thick sample in a parallelized fashion, theoretically achieving a 48-fold increase in imaging speed, compared to a single microscope of the same magnification. With this significant speed increase, we demonstrate applications of our new microscope for three potentially new cytopathology workflows: 1) for remote 3D whole-slide viewing via a custom-designed interface, 2) for remote assessment of thyroid FNA slide adequacy, and 3) to detect and predict the presence of adenocarcinoma, the most common form of lung cancer.
Results
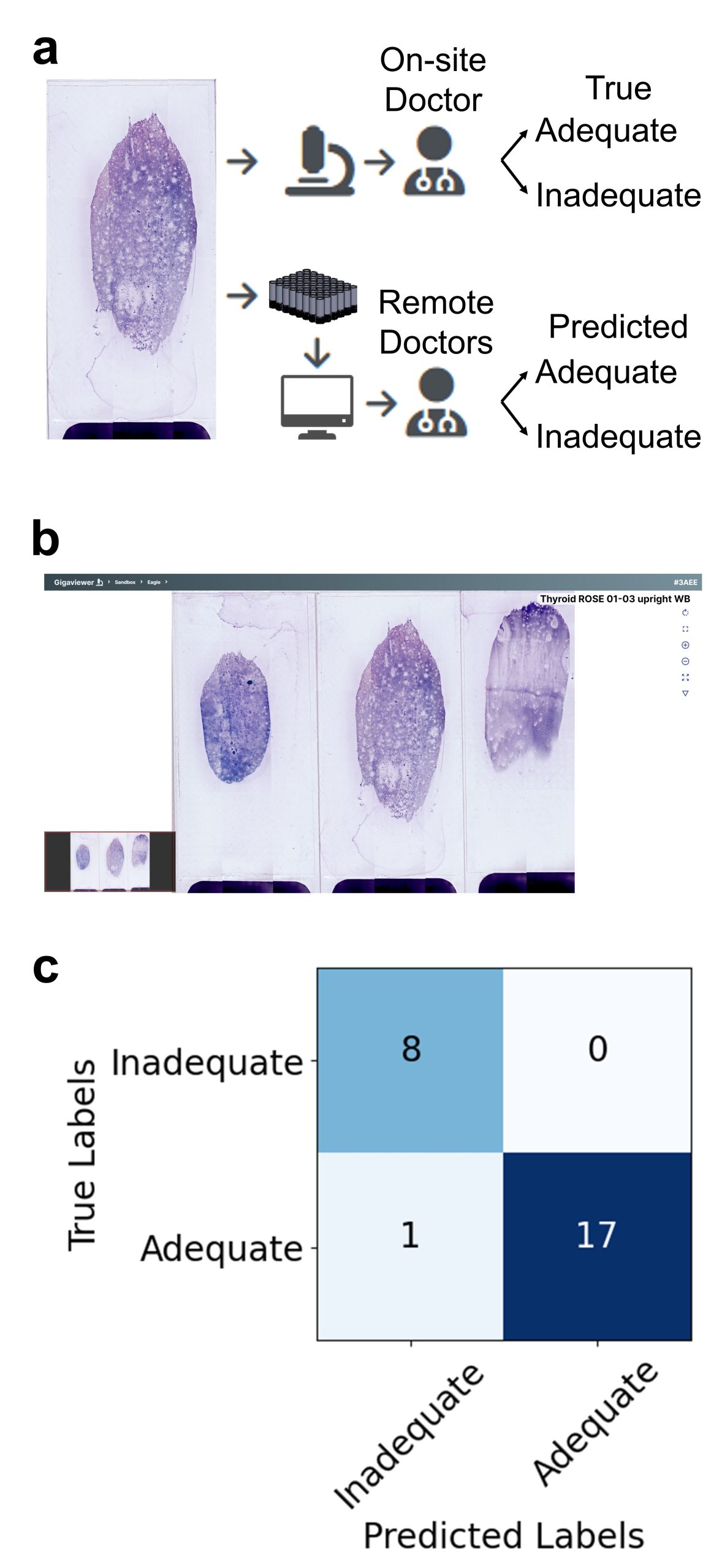
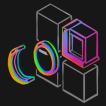
Fig 3: Evaluation of thyroid specimen adequacy. (a) One pathologist conducted the adequacy assessment using a standard microscope (True Labels). Three more pathologists used the MCAS and Gigaviewer to remotely conduct the assessment (Predicted Labels). (b) Gigaviewer interface, which is a customized viewer for remote viewing. (c) Assessments for each slide from the three pathologists are combined using majority vote, and the results are then compared to evaluate the consistency between both methods.
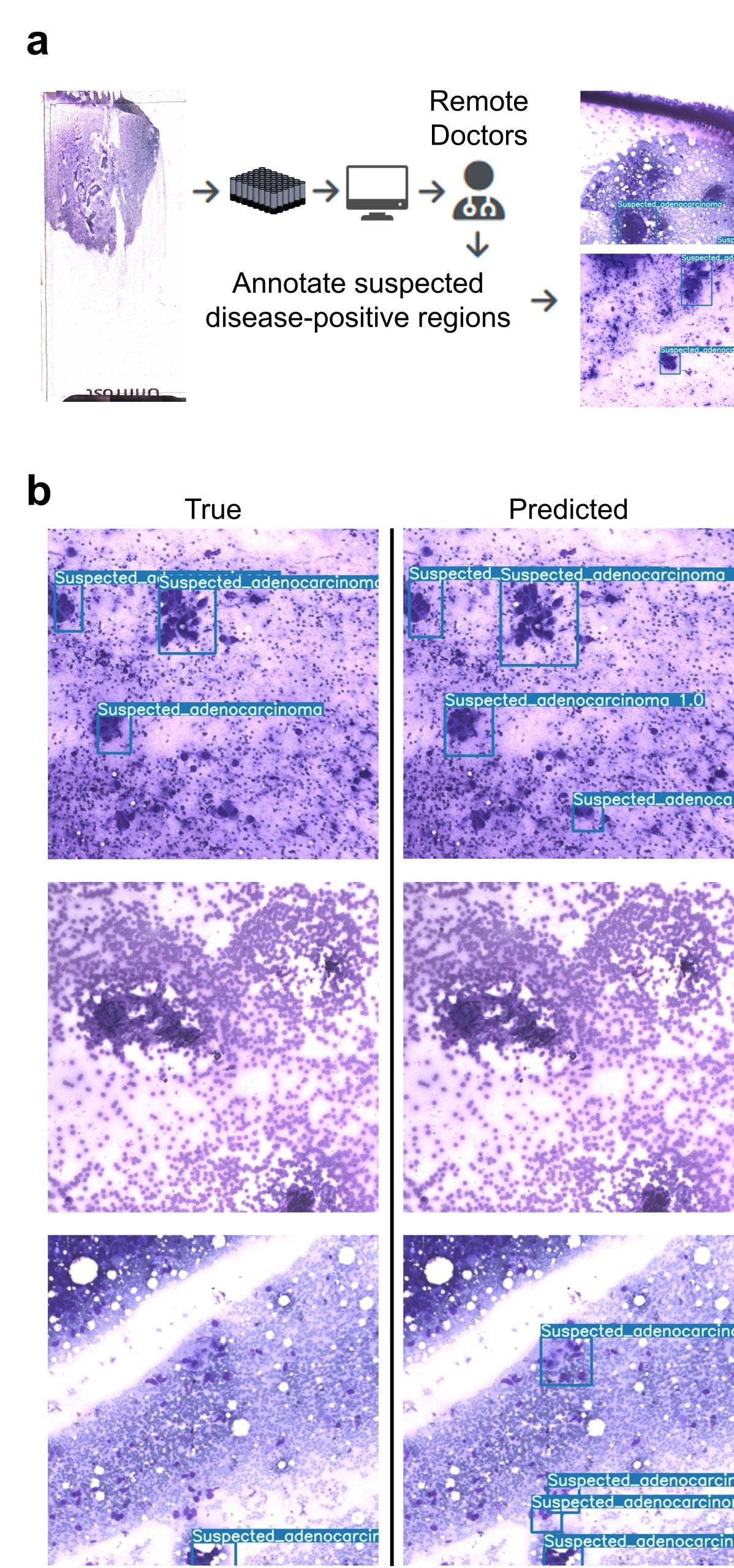
Fig 4: Detection of suspected adenocarcinoma in lung specimens using YOLOv7. (a) Pathologists annotate cancerous regions on MCAS images. (b) Machine learning results locate areas of suspected adenocarcinoma, marked as blue boxes, using a supervised learning algorithm. The trained model achieves a recall of 0.73.
Fig 5: Implementation of multiple instance learning (MIL) for accurate slide-level automated diagnosis. (a) Clinicians assigns each slide a category at the slide level using standard microscope evaluation. (b-c) The MCAS scans slides, dividing each unique camera’s FOV into 9 sub-patches to generate a `bag’ dataset. (d) Performance of MIL slide-level classification (cancer-positive vs. negative) with four-fold cross-validation.