scatterBrains: an open database of human head models and companion optode locations for realistic Monte Carlo photon simulations
Melissa M. Wu1,2, Roarke Horstmeyer1, Stefan A. Carp2
1Department of Biomedical Engineering, Duke University, Durham NC, USA.
2 Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, USA.
Abstract
Monte Carlo (MC) simulations are currently the gold standard in the near-infrared and diffuse correlation spectroscopy (NIRS/DCS) communities for generating light transport paths through tissue. However, realistic and diverse models that capture complex tissue layers are not easily available to all; moreover, manually placing optodes on such models can be tedious and time consuming. Overcoming such limitations would help enable the adoption of representative models for basic simulations, as well as the use of these models for large-scale simulations, e.g., for training machine learning algorithms.
In this work, we provide the NIRS/DCS communities with an open-source, user-friendly database of morphologically and optically realistic head models, as well as a succinct software pipeline to prepare these models for mesh-based Monte Carlo simulations of light transport. The models, head surface locations, and all associated code are freely available under the [scatterBrains project on Github](https://github.com/wumelissa/scatterBrains). We hope this will make MRI-based head models and virtual optode placement easily accessible to all. Contributions to the database are welcome and encouraged.
Database description
For each subject folder in the database, the following are included:
1. Mesh-based model of the subject head, segmented as described below
2. Volume-based model of the subject head, segmented as described below
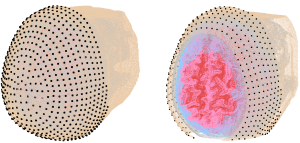
3. Companion surface locations (~800) to aid automatic optode placement, as shown below
4. Surface meshes of the subject head for user visualization
The subject database primarily consists of sixteen MRI-based head models. The MRI scans were acquired from healthy adult volunteers from a previous study, and each volume was segmented into five tissue layers representing the scalp, skull, cerebrospinal fluid, grey, and white matter. Models were then manually reviewed and polished before being converted to tetrahedral meshes.
To assist automatic optode placement, 805 predefined scalp locations are provided for each model, comprising full head coverage. Visualization of the scalp locations for an example subject are shown below.
Other functionality and usage
scatterBrains provides code to write the optode locations generated in the previous section as well as other simulation parameters (e.g. optical properties for each tissue layer) directly to an input file which can be passed to Monte Carlo simulations. After running the simulation, users can process the photon trajectory results as they wish. Additional example code for post-processing DCS is provided.
Contributions to the database (additional models or analysis code) are encouraged. Code for simulating and processing speckle contrast optical spectroscopy data is also available in the repository in a supplemental folder.
For questions or concerns please contact scatterbrains@googlegroups.com