Our review on Digital staining in optical microscopy using deep learning
________________________________________
Lucas Kreiss1,2, Shaowei Jiang3 , Xiang Li4, Shiqi Xu1, Kevin C. Zhou1,5, Kyung Chul Lee1,6,
Alexander Mühlberg2, Kanghyun Kim1, Amey Chaware1, Michael Ando7, Laura Barisoni8, Seung Ah Lee6, Guoan Zheng3, Kyle J. Lafata4, Oliver Friedrich 2 and Roarke Horstmeyer1
1 Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA
2 Institute of Medical Biotechnology, Friedrich-Alexander University (FAU), Erlangen, Germany
3 Department of Biomedical Engineering, University of Connecticut, Mansfield, Connecticut, USA
4 Department of Radiation Physics, Duke University, Durham, NC 27708, USA
5 Department of Electrical Engineering & Computer Sciences, University of California, Berkeley, CA, USA
6 School of Electrical & Electronic Engineering, Yonsei University, Seoul 03722, Republic of Korea
7 Google, Inc., Mountain View, CA 94043, USA
8 Department of Pathology, Duke University, Durham, NC 27708, USA
Link to paper
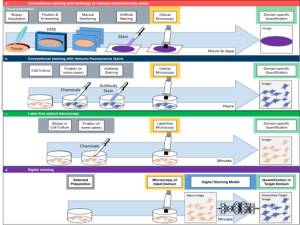
Fig 1: Basic principle of Digital staining. a) Conventional staining of 3D tissue samples requires a time-demanding and cumbersome procedure of biopsy acquisition, formalin-fixed paraffin-embedding (FFPE), manual sectioning, dehydration and artificial staining. These prepared tissue slices are then imaged by optical microscopes and the obtained images are quantified (e.g., via histopathology scoring by experienced experts). b) Staining of cell cultures is conventionally based on antibody reactions with immuno-fluorescence (IF) stains. This process does not require embedding, sectioning and dehydration, and can even compatible with live cell imaging. However, the image quantification is still specific to the applied staining (e.g., nuclei staining for segmentation of nuclei). c) Label-free optical technologies exploit the natural contrast of biomedical samples, without relying on artificial stainings. Although this omits the need for extensive sample preparation, the quantification is bound to the specific type of optical contrast that was used (e.g., dry mass approximation in quantitative phase imaging). d) Digital staining can combine the advantages of an experimentally more practical imaging technique, with the high specificity of a thorough but cumbersome staining approach. Thus, DS can be used to digitally enhance label-free optical microscopy (e.g., generation of IF images based on white light microscopy) or to perform stain-to-stain translation (e.g., generation of specific IHC staining based on already available H&E stainings)
Abstract
________________________________________
Until recently, conventional biochemical staining had the undisputed status as well-established benchmark for most biomedical problems related to clinical diagnostics, fundamental research and biotechnology. Despite this role as gold-standard, staining protocols face several challenges, such as a need for extensive, manual processing of samples, substantial time delays, altered tissue homeostasis, limited choice of contrast agents, 2D imaging instead of 3D tomography and many more. Label-free optical technologies, on the other hand, do not rely on exogenous and artificial markers, by exploiting intrinsic optical contrast mechanisms, where the specificity is typically less obvious to the human observer. Over the past few years, digital staining has emerged as a promising concept to use modern deep learning for the translation from optical contrast to established biochemical contrast of actual stainings. In this review article, we provide an in-depth analysis of the current state-of-the-art in this field, suggest methods of good practice, identify pitfalls and challenges and postulate promising advances towards potential future implementations and applications.
Overview
________________________________________
Digital staining is an emerging concept in the field of computational microscopy that can digitally augment microscopy images by transferring the contrast of input images into a target domain (see Fig. 1D). Implementation of digital models is most often based on machine learning algorithms that are trained on pairs of input and target images. In a nutshell, these ML models then learn to link characteristic features in structure and contrast from one input domain (most often a label-free image) with those of the target domain (most often images from staining with well known molecular specificity). Thereby, digital staining very elegantly bypasses two obstacles: (i) during the development and training of computational models, digital staining omits the need of manual annotations of ground truth data, by obtaining the ground truth annotations from specific stainings and (ii) upon deployment, the inference with a trained model can then circumvent the time-consuming and tedious procedure of actual sample preparation, including sectioning and staining. Thus, digital staining can be viewed as a holistic concurrence of biology, optical microscopy and computational modeling.
In this review article, we explain the general workflow of digital staining, present the most common modes of operation and categorize a great number of publications along different aspects. For instance, we grouped those publications according to the use of input images that are digitally stained and according to the target stain that the digital staining model is trained for. The figure below shows a quantitative analysis of input and target combinations as well as six examples.
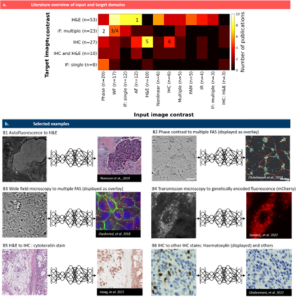
Fig. 2 Pairings of input and target contrast. a) Target image contrast is plotted against the input contrast, the number of publications in each combination is color-coded. Selected examples in (b) are indicated by numbers in (a): (B1) a translation of autofluorescence images from tissue slides to H&E images by Rivenson et al. [20], re-use permitted and licensed by Springer Nature. (B2) translation of phase contrast images of human neuron cells to specific fluorescence images (DAPI, anti-MAP2 and anti-neurofilament). Data available at https://github.com/google/in-silico-labeling from Ref. [21], re-use was permitted and licensed by Elsevier and Copyright Clearance Center. (B3) translation of bright field images of cells to multiple fluorescence stains by Ounkomol et al. [22]. Images are publicly available at https://downloads.allencell.org/publication-data/label-free-prediction/index.html, re-use was permitted and licensed by Springer Nature. (B4) translation of bright field images of living cells to genetically encoded mitochondria markers by Somani et al. [25]. Images are publicly available at https://doi.org/10.18710/11LLTW [35], re-use licensed under CC0 1.0. (B5) stain-to-stain translation of H&E images into cytokeratin stain by Hong et al. [36], Images are publicly available at https://github.com/YiyuHong/ck_virtual_staining_paper , re-use licensed under CC BY 4.0. (B6) stain-to-stain translation of IHC images into different IHC images by Ghahremani et al. [37]. Images are publicly available at https://zenodo.org/record/4751737#.YV379XVKhH4 , re-use permitted and licensed by Springer Nature. IHC = immuno-histochemcial stain, IF = immuno-fluorescence stain, WF = wide field (white light illumination), AF = autofluorescence, PAM = photo-acoustic microscopy, IR = infra-red.
Advantages
________________________________________
Similar to the previous “virtual histology” for histology images of tissue sections [18], digital staining has many advantages over actual staining, like a reduced time to perform staining, minimal manual labor, minimal stain variability, less hazardous waste composition of tissue fixatives, preservatives and staining dyes, no tissue disruption of the actual sample, no restrains to use multiple stains on a single slide, the chance to perform stain-to-stain transformation and a reduced chance for technical failures. With the extension to apply this concept to cell cultures, digital staining offers additional advantages, such as an identification of functional stages without the biochemical binding of actual antibody stains, that could otherwise interfere with biological homeostasis and impact motion, growths or other aspects of relevance.
Good scientific practice for digital staining
________________________________________
Besides an in-depth analysis and explanation of many different aspects of digital staining, and the identification of its main advantages, we put together a list of what we think are the main methods of good scientific practice, when developing digital staining.
• General feasibility: As with most ML problems, one should consider first, whether the information content in the data, i.e., the input domain, is believed to be sufficient for the given task. More specifically, a good first question is if the general information in the input images is correlated with the one in the target domain. For instance, it might seem unfeasible to digitally stain cell nuclei (target) from images that only contain fluorescence of a membrane marker as input, if no additional information was used. On the other hand, it would seem quite feasible to perform digital staining of nuclei and membrane markers based on phase contrast images, as the contrast in optical phase is high for both nuclei and membranes. If a paired data set is already available, we suggest to test the general feasibility first by developing a model for simpler tasks, such as patch classification, object detection, or semantic segmentation.
• Report uncertainties: One of the main short-comings of the current state-of-the-art for digital staining is that fundamental uncertainties in input and in target data are usually not reported. As discussed in greater detail in the full text of our review, DS is a holistic approach that involves the entire pipeline of biology, imaging and ML. Simply reporting a performance metric of the ML task, is therefore insufficient, as those metrics assume a perfect ground-truth. However, target data from actual biochemical staining are always affected by the specificity of the molecular marker as fundamental uncertainty in the ‘ground truth’. Similarly, imaging of inputs and targets is subject to the specific contrast mechanism, resolution and SNR of the respective imaging technology. Thus, we propose that digital staining should always be embedded in the context of input and target uncertainties of the actual stain as well as SNR of the imaging process to allow a fair evaluation of its performance.
• Generalizability: there is a generalization gap in deep learning and digital pathology, which also applies to digital staining. DS can often be very hardware-specific and can be prone to over-fitting. Therefore, it is essential to discuss generalizability. Ideally, one should take a hardware-agnostic approach when testing a DS pipeline. It is recommended to validate and test a system across different imaging systems and/or different tissue types to evaluate if it generalizes well. This can further be extended to evaluate the generalizability across different experimenters, different staining methods or different data sets.
• Choice of the right loss function: After the selection of input and target technologies (which might be predefined for a given problem), the choice of the loss function is important. Different loss functions can emphasize different aspects of the image-to-image regression task, e.g., high-level structural errors (SSIM), absolute errors at the pixel level (peak signal-to-noise ratios – PSNR), brightness and color (MAE) or custom loss functions (see section on Loss functions for more details).
• Image inspection and decision visualization: Besides the mere reporting of loss curves and performance metrics, it is indispensable to visually inspect and report the actual target and prediction images. Although the above mentioned loss functions are suited for training and quantitative performance comparison, some can be ill-suited to detect hallucinations [128], artifacts or other localized prediction errors in the images. Moreover, decision visualization, like occlusion maps, Shapley values or perturbation studies can inform the researcher about features that are particularly important to the learning process. This cannot only support de-bugging during the development of DS, but it can also offer valuable scientific feedback e.g., to understand which parts of an input image are particularly relevant to predict a certain target.
• Interpretability: similar to the point above, ML models can often lack interpretability, which prevents identification of biases and can thereby reduce generalizability. Interpretability is especially relevant for digital staining to prevent false hallucinations from overfitting. A good rule of thumb is that simpler models with a smaller number of parameters are more interpretable. Furthermore, it is preferred to create more interpretable models from the beginning instead of post-hoc explanations of complicated models [163].
• Availability of code & data: Whenever possible, we recommend to make code and data available to other researchers, according to the FAIR principle (Findability, Accessibility, Interoperability, and Reuse of digital assets). This enhances trustworthiness and transparency of the general scientific procedure and further enables other researchers to test new approaches, especially since good data sets of paired images might be a bottleneck for many ML researchers.
Acknowledgment
________________________________________
This work is part of a project that is funded by a Global fellowship of EU HORIZON 2022 program. Find out more details on this project here!
References
________________________________________
All references in this short summary relate to the numbering in our full review manuscript:
18. Pillar N, Ozcan A. Virtual tissue staining in pathology using machine learning. Expert Rev Mol Diagn. 2022;22(11):987–9
20. Rivenson Y, Wang HD, Wei ZS, de Haan K, Zhang YB, Wu YC, et al. Virtual histological staining of unlabeled tissue-autofluorescence images via deep learning. Nat Biomed Eng. 2019;3(6):466–77.
21. Christiansen EM, Yang SJ, Ando DM, Javaherian A, Skibinski G, Lipnick S, et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell. 2018;173(3):792.
22. Ounkomol C, Seshamani S, Maleckar MM, Collman F, Johnson GR. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat Methods. 2018;15(11):917.
25. Somani A, Sekh AA, Opstad IS, Birgisdottir ÅB, Myrmel T, Ahluwalia BS, et al. Virtual labeling of mitochondria in living cells using correlative imaging and physics-guided deep learning. Biomed Opt Express. 2022;13(10):5495–516
35. Opstad I. Data set: Fluorescence microscopy videos of mitochondria in H9c2 cardiomyoblasts. DataverseNO. 2023
36. Hong Y, Heo YJ, Kim B, Lee D, Ahn S, Ha SY, et al. Deep learning-based virtual cytokeratin staining of gastric carcinomas to measure tumor-stroma ratio. Sci Rep. 2021;11(1).
37. Ghahremani P, Li Y, Kaufman A, Vanguri R, Greenwald N, Angelo M, et al. Deep learning-inferred multiplex immunofluorescence for immunohistochemical image quantification. Nat Mach Intell. 2022;4(4):401–12
128. Cohen JP, Luck M, Honari S. Distribution matching losses can hallucinate features in medical image translation. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science. vol. 11070. Cham: Springer. 2018. p. 529–3
163. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1(5):206–15