Deep Optical Blood Analysis: COVID-19 Detection as a Case Study in Next Generation Blood Screening
medRxiv(2021)
, , , , , , , , , , ,
Code Paper
Abstract
Visual examination of cell morphology within a peripheral blood smear, is a common method for a diagnosing a wide variety of diseases. In this project, we propose a novel machine learning approach that offers a powerful and scalable means to analyse peripheral blood smears. We utilise a multiple instance learning based approach to understand the morphological impact of COVID-19 on the blood cells across the various cell types, which is still not well understood. This quantification methodology enables high-throughput extraction of the high-resolution morphological information from the peripheral blood smears, resulting into an automatic diagnostics tool. Moreover, our study integrates diagnostic and image information from across 236 patients to establish a significant link between blood and a patient’s COVID-19 infection status. Our results also corroborate well with the related hematological findings that examine the impact of COVID-19 on the blood cell morphology. We report a high diagnostic efficacy; with an accuracy of 79% and ROC-AUC of 0.90.

Introduction
Recent clinical findings suggest a complex set of interactions between COVID-19 and blood, especially morphological changes, that lead to significant mortality and morbidity among the infected patients. However, our limited understanding of such an impact has impeded the development of an effective blood-based screening tools
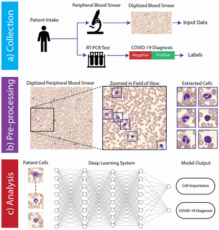
We present a new way of analysing blood, which we term Deep Optical Blood Analysis (DOBA), allows for an entirely data-driven analysis of blood using only patient-level information, without pre-defining features of interest or label individual cells within particular categories. “DOBA uses deep learning to develop a mapping between images from a patient’s PBS and their condition. In a typical digital PBS scan, images of hundreds of white and red blood cells are captured per-patient. It is therefore desirable to examine each image in detail, without requiring labels on the individual images. To accomplish this, we adopted a Multiple Instance Learning(MIL)1 technique to link a patient’s COVID-19 diagnosis (obtained with a standard PCR-RT laboratory test) to their blood image data.”
Result
Study Design
We investigated the diagnostic potential of PBS images for COVID-19 infection through a partnership with the Duke University Medical Center. Over a five month period (April 2020 – August 2020) we collected digital PBS image data from 236 patients, 53% of whom tested positive for COVID-19 by a separately administered PCR test. No other patient information was collected for this cohort. In addition, we collected PBS image data from 40 additional patients admitted to the medical intensive care unit who presented with acute respiratory illness, but were confirmed to COVID-19 negative using the same PCR testing method. We denote these two cohorts of patients as the Standard and Challengegroups throughout this work.
PBS image data was collected using a clinically approved digital slide scanner (Cellavision DM9600), which uses an oil immersion objective lens to capture multiple high resolution images per-patient centered upon stained (Wright-Giemsa) white blood cells (WBCs), with an average of 130 images captured per-patient. To preserve patient privacy, no additional data, such as demographic information, was collected.
The standard cohort included 236 patients, 125 of whom tested positive for COVID-19. all performance metrics are reported as the average test-set performance across all six folds, where multiple independent models were trained from scratch exclusively for individual folds. This strategy enabled us to test our system on all available data while isolating the test data during the training process.
Machine Learning System and Results

Overview of hybrid machine learning system. The SIL branch processes each image from a patient PBS scan individually. The outputs are then aggregated to produce a single prediction (using the median of the single image predictions). The MIL branch collectively analyzes all of a patient’s images simultaneously, producing one prediction per patient. This is accomplished by first extracting learned per-image features, and then feeding those features into an attention module. The attention module assigns weights (summing to one) to each learned feature. The weights are used to compute a weighted sum across image features, the result of which is passed into a classification module to produce the MIL patient prediction. These two strategies are combined through ensembling, where the outputs of each branch are averaged to produce the final outcome
 Performance of COVID-19 diagnosis from blood cell morphology analysis as measured by the receiver operator characteristic (ROC). Classification accuracy was 79% and 82% for the standard and challenge cohorts respectively. a) Results reported are the average across the entire dataset, k-fold cross validation was used to maintain independence between the training and test sets. b) COVID-19 positive patients were randomly selected from the standard cohort to counterbalance the COVID-19 negative patients from the challenge cohort (ROC-AUC calculation requires both positive and negative examples).
Performance of COVID-19 diagnosis from blood cell morphology analysis as measured by the receiver operator characteristic (ROC). Classification accuracy was 79% and 82% for the standard and challenge cohorts respectively. a) Results reported are the average across the entire dataset, k-fold cross validation was used to maintain independence between the training and test sets. b) COVID-19 positive patients were randomly selected from the standard cohort to counterbalance the COVID-19 negative patients from the challenge cohort (ROC-AUC calculation requires both positive and negative examples).